Cryogenic electron tomography (cryo-ET) is an imaging technique that visualizes biological molecules and intracellular structures in their native 3D environments at nanometer resolution. In cryo-ET, a three-dimensional sample is observed by tilting it around a fixed axis while capturing two-dimensional projections at known viewing directions using the transmission electron microscope (TEM).
End-to-end localized deep learning for Cryo-ET
Cryo-electron tomography (cryo-ET) enables 3D visualization of cellular environments. Accurate reconstruction of high-resolution volumes is complicated by the very low signal-to-noise ratio and a restricted range of sample tilts, creating a missing wedge of Fourier information. Recent selfsupervised deep learning approaches, which post-process initial reconstructions done by filtered backprojection (FBP), have significantly improved reconstruction quality, but they are computationally expensive, demand large memory, and require retraining for each new dataset. End-to-end supervised learning is an appealing alternative but is impeded by the lack of ground truth and the large memory demands of high-resolution volumetric data. Training on synthetic data often leads to overfitting and poor generalization to real data, and, to date, no general end-to-end deep learning reconstructors exist for cryo-ET. In this work, we introduce CryoLithe, a local, memory-efficient reconstruction network that directly estimates the volume from an aligned tilt-series, overcoming the suboptimal FBP. We demonstrate that leveraging transform-domain locality makes our network robust to distribution shifts, enabling effective supervised training and giving excellent results on real data—without retraining or fine-tuning
Methodology
Cryolithe is a localized, end-to-end supervised framework for cryo-ET reconstructions. To enable data-efficient training and ensure robustness to out-of-distribution shifts (experimentally), we design the network to operate on small patches of fixed size. Our method first extracts 2D patches using the geometric relationship defined by the forward (physical) model and then reconstructs the volume in a voxel-wise manner. As a result, it requires only a few volumes for training and can recover denoised reconstructions across a wide variety of datasets.
Patch Extraction
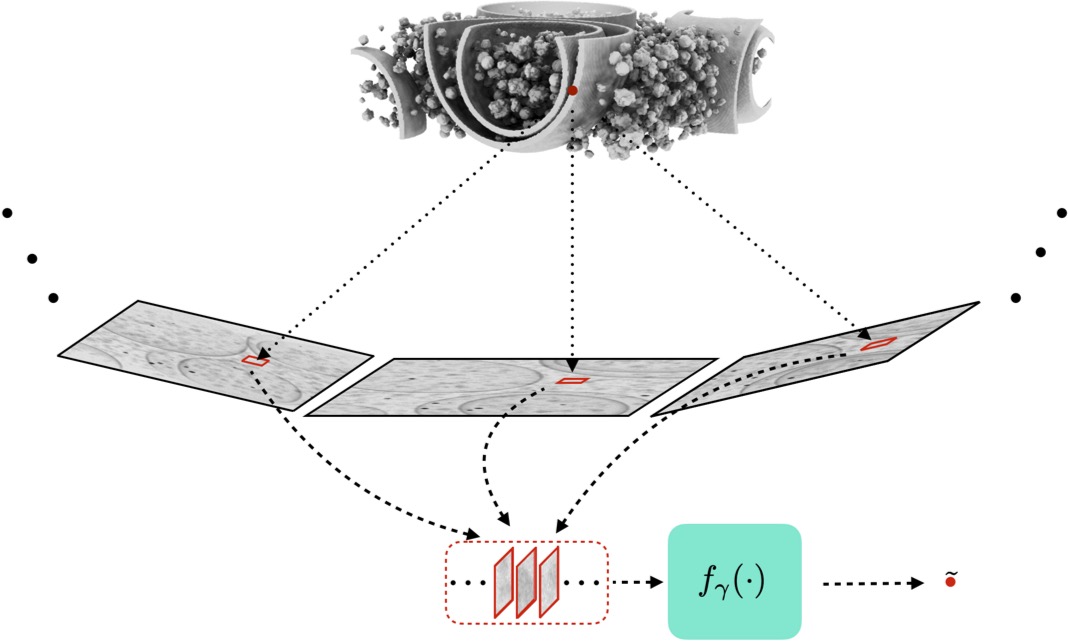
The patch extract process is based on the Radon (X-ray) transform. As shown in the below figure, for each voxel, we identify the corresponding pixel in the projection that is influenced by that voxel. This mapping depends on the beam direction and the position the voxel's position. We extract a small patch around the affected pixel from all measured projections. These projections are fed into the network to recover the voxels value.
Figure: Overview of the patch extractor. The figure highlights the part of the projections where the volume at a particular location (red dot) is observed. CryoLithe extracts a local patch around these locations, which is processed by a neural network.
Note: In practice, we use ramp-filtered projections instead of raw projections. This helps the network with the global context.
Architecture
The network is composed of multilayer perceptrons (MLPs) with predefined depth and width. Input patches are divided into slices based on the imaging geometry. Each slice is processed by an individual MLP to produce a slice-wise estimate, represented as a feature vector of a fixed size. These feature vectors are then concatenated and passed through another MLP to produce the final voxel estimate. This design reduces the overall size of the MLPs while effectively integrating information from all slices. The architecture is inspired by the MLP-mixer, with modifications to account for the Radon transform geometry.
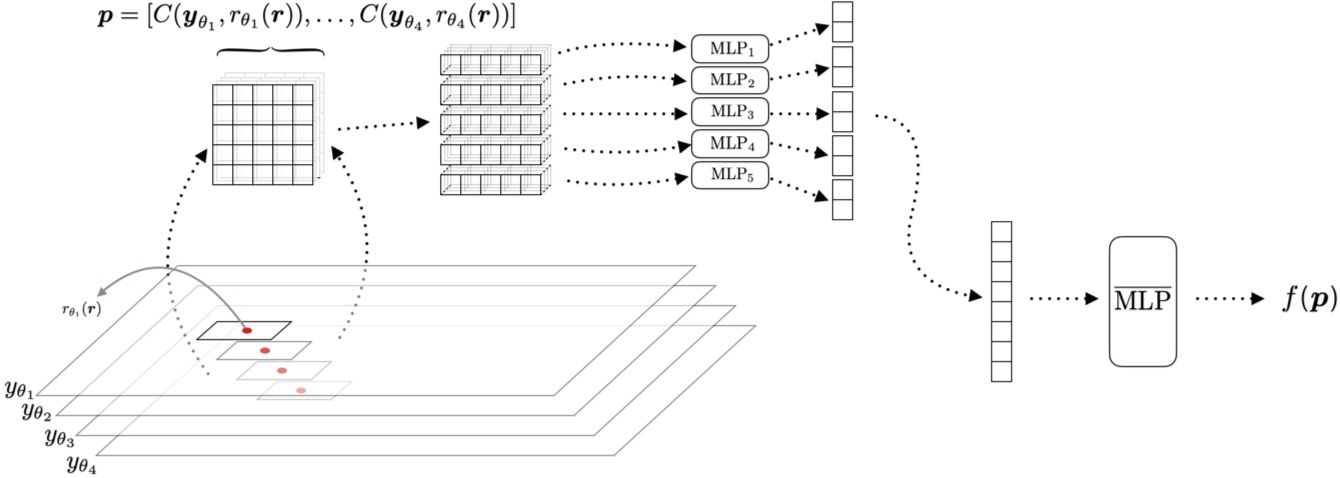
Figure: Schematic diagram of the architecture. The patches are grouped along the slices based on the axis of the tilt series. These slices are fed into the corresponding MLPs (MLPi) to obtain a feature vector, which is used by a combination MLP (MLP) to estimate the volume at a particular location r. While the architecture appears slicewise for computational reasons, it is fundamentally a 3D architecture that exploits the 3D correlation structure of the measurement data to recover the volume.
Training
Due to the lack of ground truth volums. we use outputs from self-supervised networks as the references. These networks are trained on individual projections, which reduces the liklihood of hallucinated features in the reconstructions. We then carefully select reference volumes based on how well they have been denoised debiused by the self-supervised networks. This selection is done visually by discarding reconstructions with arefacts or those that appear less denosied compared to standard filtered back-projection reconstructions.
Results
Cryolithe is trained using reconstruction obtained from cryo-CARE + IsoNet. As a result, its outputs are generally comparable to those of self-supervised methods-sometimes achieving better quality and, in other cases, slightly lower fidelity. However, we observe that Cryolithe avoids many of the artifacts that commonly appear in self-supervised reconstructions for certain volumes. We believe this difference may be partly due to the non-convex training landscape of self-supervised methods like IsoNet. In cases where slight instabilities are present, the training process—which involves repeatedly applying the network over several iterations—can amplify these issues, potentially leading to visible artifacts. While careful tuning of hyperparameters might mitigate such effects, the inherent sensitivity of the self-supervised training process can still make it less stable across datasets. In contrast, there is no retraining in Cryolithe, thus no large-scale instability in the output.
Since it does not require retraining for each dataset, it is considerably faster to apply in large-scale or high-throughput settings. This makes it particularly attractive for scenarios where time and computational resources are limited.
In the following sections, we present qualitative comparisons between Cryolithe and other methods to showcase its performance as well as the time required to obtain the volume starting from noisy reconstruction for self-supervised methods and tilt series for Cryolithe.
Runtime
We test the runtime for different methods to recover a simulated volume of size 800x 800x400. We generate 41 projections from the volume using tomosipo package. For self-supervised methods, we take into account the training time as well. We used the default parameter mentioned by the authors of the methods to train the self-supervised models.
| FBP+Cryo-CARE | FBP+IsoNet | FBP+Cryo-CARE + IsoNet | FBP+DeepDeWedge | CryoLithe | CryoLithe-wavelet |
|---|---|---|---|---|---|
| 1:14:36 | 3:58:24 | 5:13:00 | 17:03:09 | 0:23:36 | 0:03:07 |
Table 1: Run time of the different methods in hours:minutes:seconds format for processing a tilt-series of size 41 × 800 × 800 into a tomogram of size 800 × 800 × 400.
Visual comparison
We compare the reconstruction visually along with cryo-care+ isonet reconstruction. Since cryo-care requires two sets of the volumes to train in a Noise2Noise manner. We split the volume into odd and even tilts and train cryo-care.
Figure: Use the slider to compare cryo-ET image reconstructions side by side, and adjust the Z-bar to observe image layers across different depths.
How do I use it ?
To run the code on your dataset. Download the files and install the necessary libraries. The installation instructions are provided in the GitHub repo: github.com/swing-research/CryoLithe
Once installed. We provide you with several models to obtain the reconstruction. The fastest is currently the wavelet model.
We provide you with a sample .yaml file with the necessary parameters. You can change the input tilt series location and the angle file, as well as the location at which the output is to be saved. Once create you can run the following command to obtain the denoised volume:
We tested our codes in A100, RTX3090,RTX4090 GPUS.
Publications
2025
@article{kishore2025end,
title={End-to-end localized deep learning for Cryo-ET},
author={Kishore, Vinith and Debarnot, Valentin and Righetto, Ricardo D and Khorashadizadeh, AmirEhsan and Dokmani{\'c}, Ivan},
journal={arXiv preprint arXiv:2501.15246},
year={2025}
}